Abstract
Introduction
Current staging methods do not accurately predict clinical outcome of patients with chronic lymphocytic leukemia (CLL) especially in the new era of immunotherapy. Recent studies suggested that immune-related gene signature predicted survival in glioblastoma, breast cancer and colorectal cancer. However, none related studies have been elucidated in CLL. Here, we hypothesized that risk-prediction model integrated with the immune signature could serve as an effective prognostic indicator in CLL. This study aimed to identify reliable and distinct immune-associated fingerprints for robust classification and survival prediction in patients with CLL.
Methods
A total of 720 de novo CLL patients from multiple cohorts were enrolled with informed consents in the present study. LASSO Cox regression model was utilized to calculate immune-associated risk score (I-score) in R software. Principal Component Analysis (PCA) were performed to present the distribution of risk score. Moreover, the prognostic capability of the five immune-related fingerprints was demonstrated by PCR and ROC curve analysis with leave-one-out cross validation in the training set. Functional enrichment analyses of GO and KEGG in gene expression datasets were performed. Association between I-score and hallmark gene sets from the Molecular Signatures Database (MSigDB) were analyzed using GSEA software. Furthermore, preclinical experiments were conduct to examine the pathological mechanism of constitutive genes of I-score in CLL cell lines (MEC1, EHEB) and primary cells. Additionally, genomic regulatory network was displayed in Cytoscape software.
Results
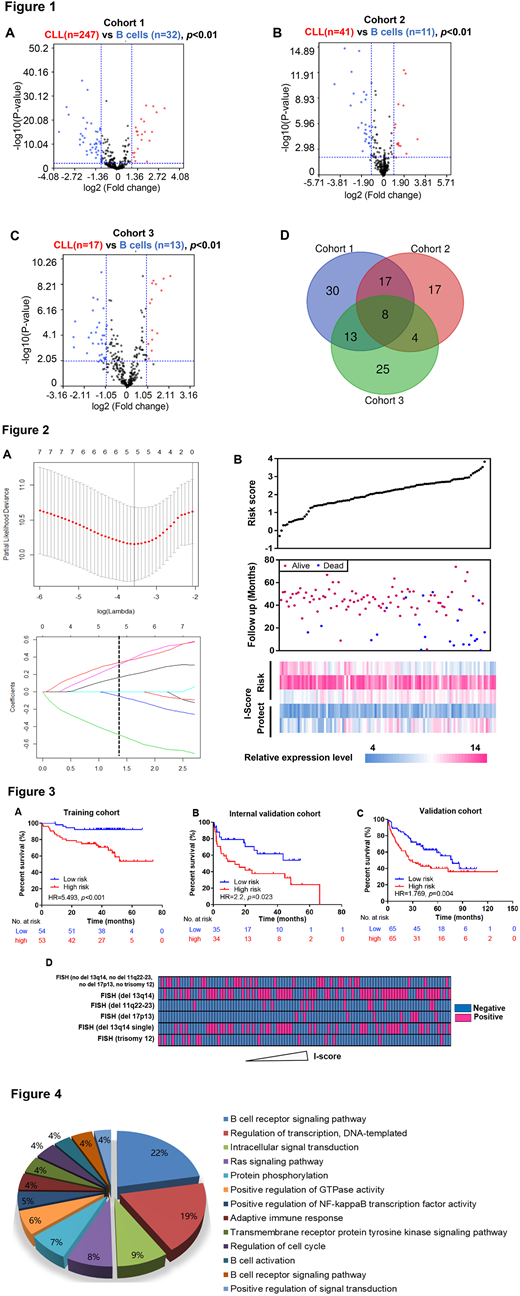
In the present study, we performed a comprehensive analysis to dissect the immune-associated fingerprints in CLL. A total of 305 clinical annotated CLL patients and 56 healthy donors with gene expression data were obtained from three independent cohorts. Two gene sets (immune system process, M13664 and immune response, M19817) were extracted from the MSigDB and combined to integrate the immune-related gene set containing 322 genes. Illustrated in the volcano plots, differentiated expressed genes of CLL cells comparing with normal B cells were calculated by limma test (|Log2Fold Change|>1, p<0.01; Figure 1A-C). Venn diagram was delineated to generate the specific CLL immune-associated gene expression panel, with 8 genes were identified (Figure 1D). PCA showed a different distribution pattern, confirming the enhanced immune phenotype in CLL.
Then, we further investigate the association of the immuno-signature with clinical outcomes of CLL patients. By Lasso Cox regression analysis, the prognostic immune-related fingerprints were identified in the training set (Figure 2A). The risk score method was established: I-score = (-0.538)*CD3D expression+(-0.077)*CD83 expression+0.364*LAX1 expression+0.191*IL2RA expression+0.362*AIM2 expression, consisting of protective genes (CD3D, CD83) and risky genes (LAX1, IL2RA and AIM2; Figure 2B).
Based on the median level of I-score as cut-off value, stratified high-risk patients were observed with significantly shorter overall survival compared with the low-risk group (Hazard Ratio, HR=5.493, p<0.001; Figure 3A). Univariate and multivariate cox regression analyses confirmed high I-score as an independent prognosis biomarker in CLL patients. To reduce disparity of diverse populations, we evaluated I-score in two independent centers of US and Germany with the same formula. The prognostic value of the immune fingerprints were corroborated in the internal validation series (HR=2.200, p=0.023; Figure 3B) and validation series (HR=1.769, p=0.004; Figure 3C). Intriguingly, we also observed the significant correlation between high I-score and 17p13 deletion (p=0.0316; Figure 3D), in accordance with patients' inferior outcome. Moreover, functional enrichment analyses of differentiated expressed genes stratified by I-score indicated that immune related BCR signaling pathway contributed to pathogenesis of CLL (Figure 4).
Conclusion
To date, our study provides evidence for the first time that distinct immuno-related fingerprints predict survival in CLL. I-score is demonstrated as an efficient classification tool and robust method for prognosis evaluation, greatly facilitating risk stratification and individualized management of CLL patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.